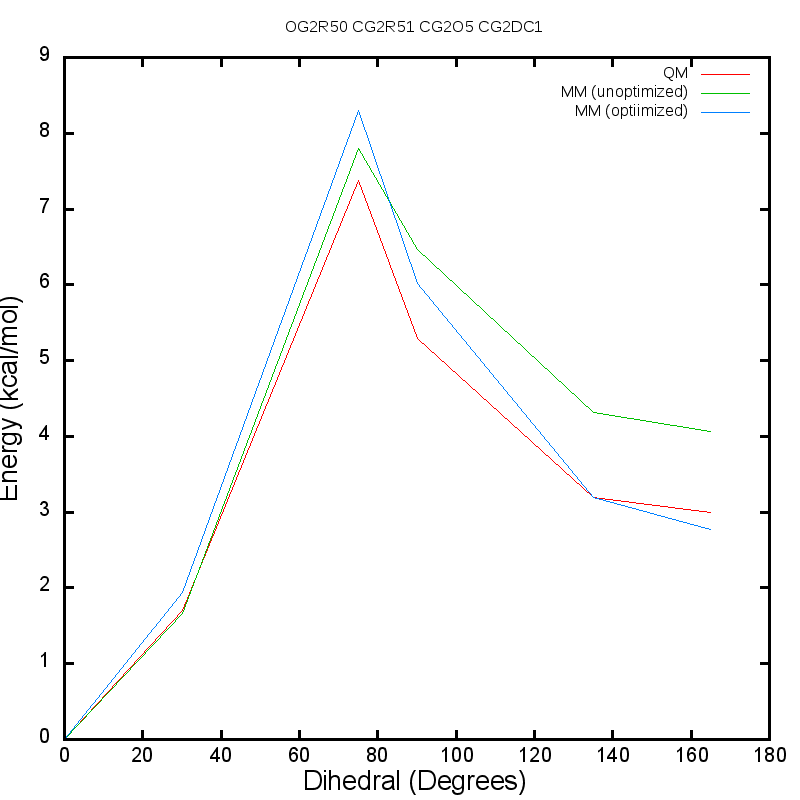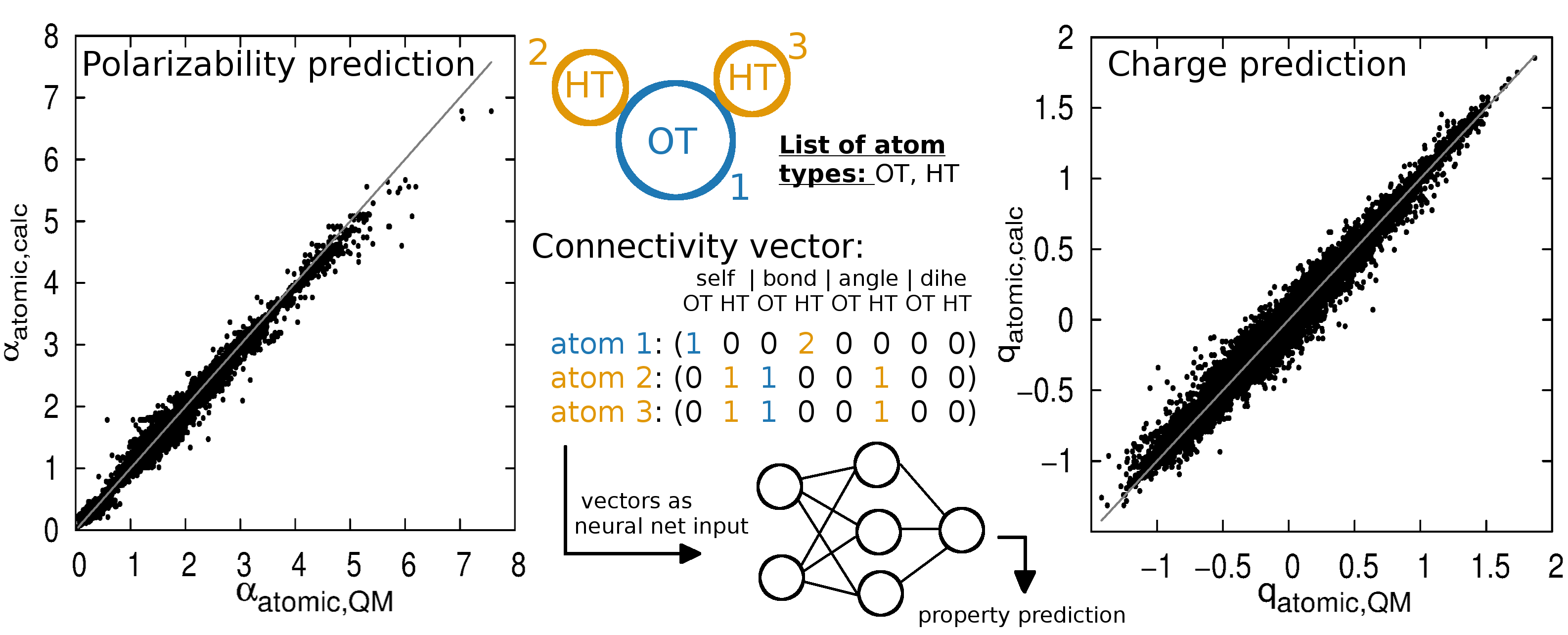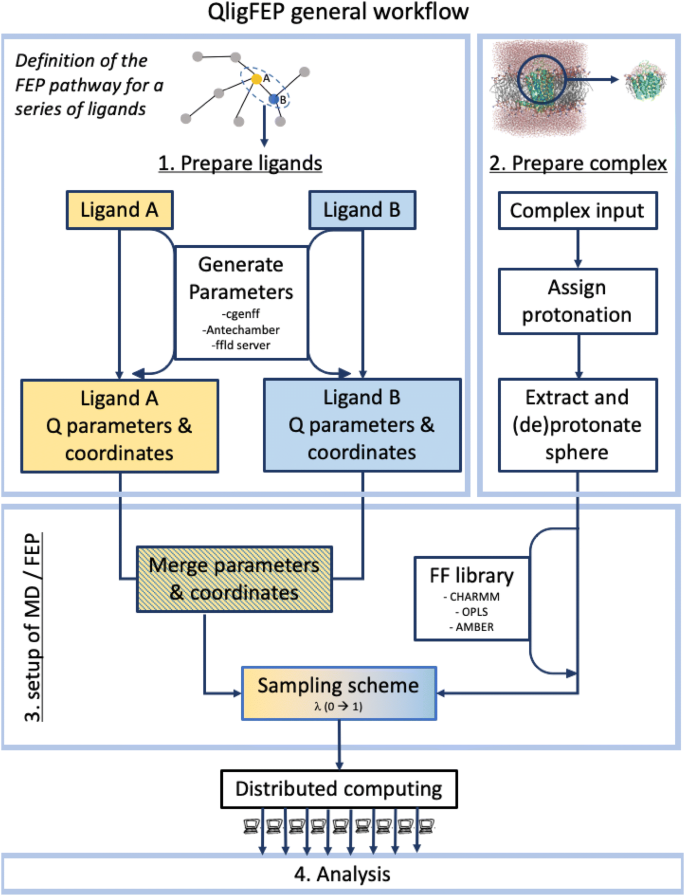
QligFEP: an automated workflow for small molecule free energy calculations in Q | Journal of Cheminformatics | Full Text
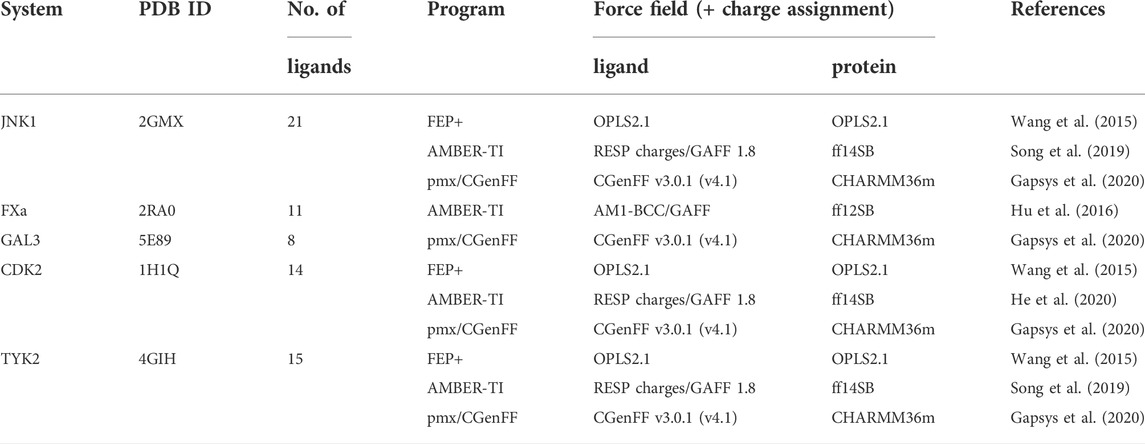
Frontiers | Relative binding free energy calculations with transformato: A molecular dynamics engine-independent tool
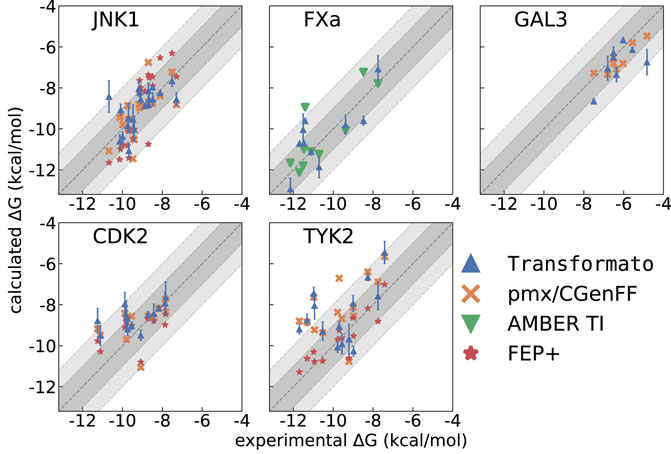
Frontiers | Relative binding free energy calculations with transformato: A molecular dynamics engine-independent tool
GitHub - jandom/paramchem: Python scrapper for paramche/cgenff server, small molecule parametrization

Problem in file (Broken structure ini.pdb) generated using CGenFF for proceeding in MD Simulation - User discussions - GROMACS forums
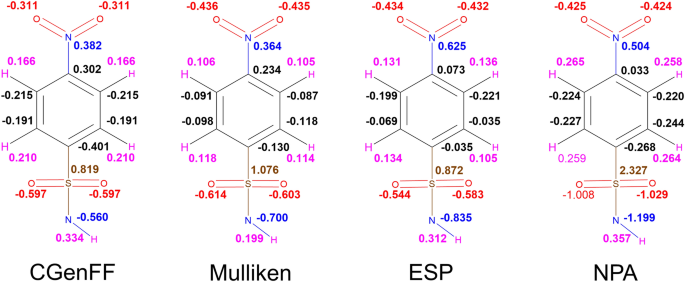
MM/GBSA prediction of relative binding affinities of carbonic anhydrase inhibitors: effect of atomic charges and comparison with Autodock4Zn | Journal of Computer-Aided Molecular Design

FFParam: Standalone package for CHARMM additive and Drude polarizable force field parametrization of small molecules - Kumar - 2020 - Journal of Computational Chemistry - Wiley Online Library

CHARMM-GUI PDB Manipulator: Various PDB Structural Modifications for Biomolecular Modeling and Simulation - ScienceDirect

CHARMM force field parameters for 2′-hydroxybiphenyl-2-sulfinate, 2-hydroxybiphenyl, and related analogs - ScienceDirect
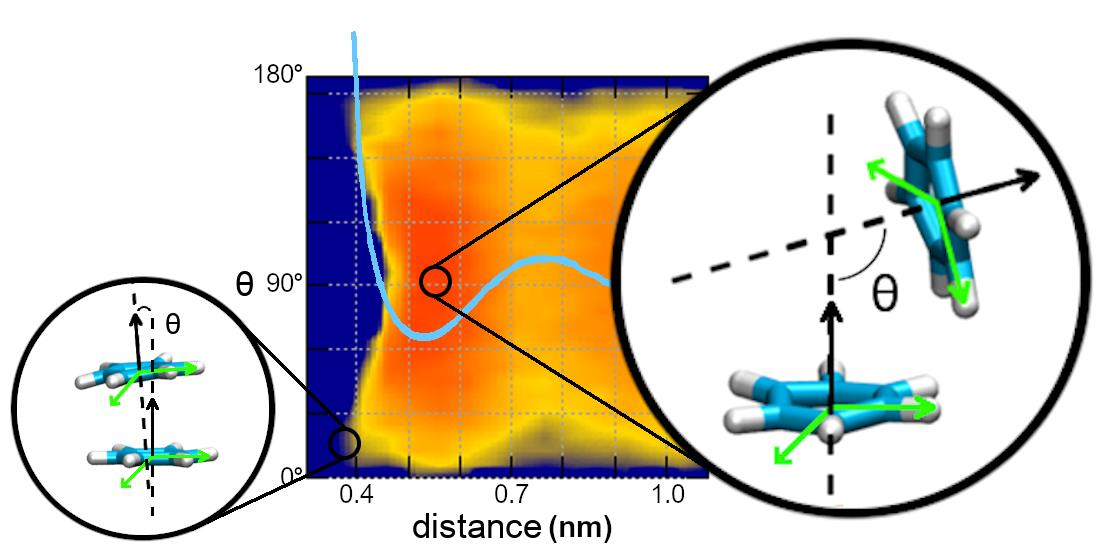
Molecules | Free Full-Text | Comparing Dimerization Free Energies and Binding Modes of Small Aromatic Molecules with Different Force Fields
molecular dynamics - Alternative to CGenFF for generating large ligand topology - Matter Modeling Stack Exchange